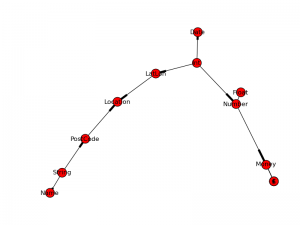
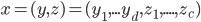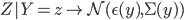As previously discussed Bayesian Belief Networks are being used within this project to perform high level type inference. To do this inference will be performed using the constructed graph.
However before inference can take place it is first necessary to train the Bayesian Network.
The graph being used for this initial test contains the following nodes:
LatLon, £, $, Int, Money, Float, Location, Number, €, String, PostCode,
Date, Name
It can be represented graphically as follows:
The labeling of data for training is implemented in two ways.
The first method uses the basic type inference system as previously described. For each piece of data each data type is considered and if the method returns true it can be determined that a positive observation for that datatype has occurred.
The second of method involves the hand labeling of data columns. When the caching system (as previously described) reads in the data it produces an additional text file that stores the column labels and allows for the columns to be labeled by hand. When the cached data is read in for processing at a later date the labels are loaded into to the data structure that contains the data.
When the training method is called it can then look to see if there is a label for this column and hence for every piece of data in that column mark a positive observation for that datatype.
Storing the Evidence
Currently conditional probability tables (CPTs) are used to store the training data. This has the limitation in that only features that are boolean in nature, such as "is it raining?", can be represented. Meaning continuous data, such as quantity of rainfall, can not be represented at this time.
With this said the CPTs are represented internally in the following way.
For each row in the CPT there is the following:
[Array of dependency nodes], [Array representing the boolean values of the dependencies], [Positive Observations, Total Observations]
Intuitively it can be seen that a probability can be derived by the following,
P = Positive Observations/Total Observations
And similarly the probability of it not being a particular type can be derived by the following,
¬P = 1-Positive Observations/Total Observations
The probabilities are not stored as a float internally for efficiency reasons.
The number of rows in a CPT is equal to to 2^(# of dependency nodes). To derive all the combinations of True and False values for all of these rows is simple. One simply counts from 0 to the number of rows and converts the integer number to a binary string and then converts it to an array of True and Falses.
When the training system identifies a positive observation it must be attributed to the correct row within the CPT. To do this it traverses all the nodes linked into the current node, for each it get a True or False value which represent their state in regards to this piece of data. These are assembled into an array matching the format of that within the CPT. The CPT is then traversed until a matching array is found and hence the correct row has been found. The positive observation is then attributed to this row.
Output
Please note that 1. the training set is very limited and 2. the basic type inference is not complete so the actual values here are inaccurate but it serves to demonstrate the training system.
LatLon
Int * Present Observed
****************************
F * 0 598
T * 0 12
£
Present Observed
******************
0 610
$
Present Observed
******************
0 610
Int
Present Observed
******************
12 610
Money
£ Number $ € * Present Observed
*******************************************
F F F F * 61 610
F F F T * 0 0
F F T F * 0 0
F F T T * 0 0
F T F F * 0 0
F T F T * 0 0
F T T F * 0 0
F T T T * 0 0
T F F F * 0 0
T F F T * 0 0
T F T F * 0 0
T F T T * 0 0
T T F F * 0 0
T T F T * 0 0
T T T F * 0 0
T T T T * 0 0
Float
Present Observed
******************
73 610
Location
LatLon PostCode * Present Observed
*******************************************
F F * 0 610
F T * 0 0
T F * 0 0
T T * 0 0
Number
Int Float * Present Observed
************************************
F F * 171 537
F T * 0 61
T F * 0 0
T T * 12 12
€
Present Observed
******************
0 610
String
Present Observed
******************
610 610
PostCode
String * Present Observed
*******************************
F * 0 0
T * 0 610
Date
Int * Present Observed
****************************
F * 61 598
T * 0 12
Name
String * Present Observed
*******************************
F * 0 0
T * 0 610